Contents
 (Image reproduced from: https://blog.f1000.com/2017/02/01/f1000prime-f1000prime-faculty-launch-bioinformatics-biomedical-informatics-computational-biology/)
(Image reproduced from: https://blog.f1000.com/2017/02/01/f1000prime-f1000prime-faculty-launch-bioinformatics-biomedical-informatics-computational-biology/)
Computational Biology
Computational biology is the combined application of math, statistics and computer science to solve biology-based problems. Examples of biology problems are: genetics, evolution,cell biology, biochemistry. [1]
Introduction
Recent advancements in technology are enabling us to store an incredible amount of data. Initially, “Big Data” was perceived as a problem to be solved. In fact, we had reached a point in which we were able to store too much data without being able to make the best use of it. This drove a need for advancements in Data Science and Artificial Intelligence.
Nowadays, what was considered first as a problem, has now become an open door to a world of innovations. Big Data has enabled many research fields such as Computer Vision and Deep Learning to flourish. This made possible for machines to perform complicated decision-making tasks and to extract from raw data information hidden, until then, to the human eye.
Biology is a subject which makes wide use of biological databases to try to tackle many different challenges such as understanding the treatment for diseases and cellular function. Datasets of biological data can be created from amino-acid sequences, nucleotides, macromolecular structures and so on.
Additionally, many robotic systems and algorithms frequently used in Computer Science are inspired by biological complexes. For example, Deep Learning Neural Networks are inspired in principle by the human brain structure.
Computational Biology Algorithms
Some examples of algorithms used in computational biology are:
- Global Matching
- Local Sequence Matching
- Hidden Markov Models
- Population genetics
- Evolutionary Trees
- Gene Regulation Networks
- Chemical Equations
Global Matching (also known as the Needleman-Wunsch problem) and Local Sequence Matching (also known as the Smith-Waterman problem) makes use of our knowledge about the proteins of an organism to understand more about other organisms proteins.
Markov Models are used for modelling sequences. In these types of models, the probability of an event to happen is just dependent on its previous state (this type of model can, for instance, be used to model a DNA sequence). Hidden Markov Models (Figure 1) makes instead use of a probabilistic Finite-state machine in which, depending on the probability of the state we are in, we emit a letter and then move to the next state. The next state can possibly be equal to the original one.
 Figure 1: Hidden Markov Model [2]
Figure 1: Hidden Markov Model [2]
Population genetics tries to model evolution. To do so, it commonly makes use of the Fisher-Wright Model. This model tries to simulate what happens at the location of a gene in selection, mutation and crossover conditions.
Evolutionary trees (Figure 2) can be created based on some form of evolutionary distance. There are two main types of evolutionary trees: distance-based trees and sequence-based trees. Evolutionary trees are used to explain distances between different species.

Figure 2: Evolutionary Trees [3]
Gene regulation networks are formed thanks to the interaction of different proteins in an organism. The different proteins control’s each other and according to the nature of their interactions, the cell type is determined.
Chemical equations (Figure 3) can finally be used to describe the mechanics behind gene regulation networks. The reaction rates are dependent on the concentration of the elements in the chemical equations.
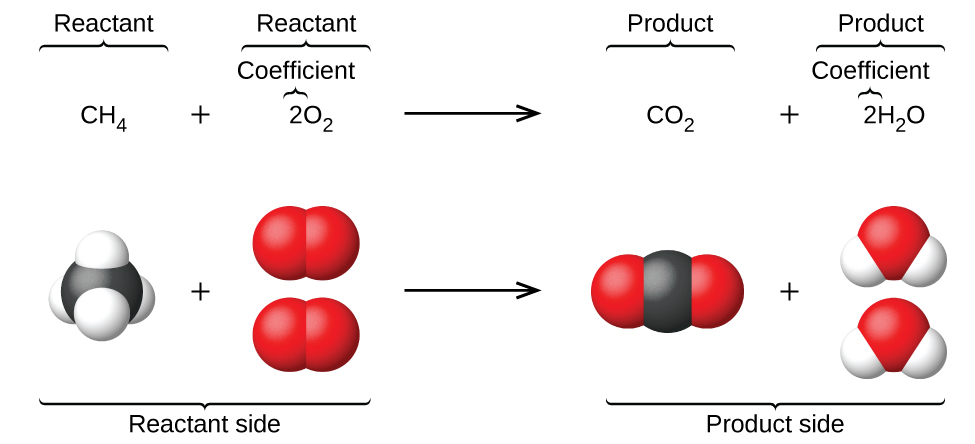 Figure 3: Chemical Equations [4]
Figure 3: Chemical Equations [4]
Machine Learning for biological prediction
Use of Machine Learning in Computational Biology is now becoming more and more important (Figure 4). Currently, applications are genomics (to study an organism’s DNA sequence), proteomics (to better understand the structure and function of different proteins) and cancer detection.
 Figure 4: Machine Learning workflow in Biological Data Analysis [5]
Figure 4: Machine Learning workflow in Biological Data Analysis [5]
Researchers demonstrated that using Convolutional Neural Networks (CNN) and Computer Vision, on cancer detection through image classification, can result in appreciable classification accuracy [6].
Several datasets are publicly available online to get started on biological data exploration, one example can be xenabrowser.net. These datasets are provided by UCSC Xena, University of California Santa Cruz.
Bibliography
[1] Computer Science Degree Hub. Accessed at: https://www.computersciencedegreehub.com/faq/what-is-computational-biology/ , May 2019.
[2] Hidden Markov Model. Accessed at: https://en.wikipedia.org/wiki/Hidden_Markov_model ,May 2019.
[3] Of BATs and APEs: An interactive tabletop game for natural history museums. Michael S. Horn et al. Accessed at: https://www.researchgate.net/publication/254004984Of_BATs_and_APEs_An_interactive _tabletop_game_for_natural_history museums/figures?lo=1, May 2019.
[4] Chemistry LibreTexts. 4.1: Chemical Reaction Equations. Accessed at: https://chem.libretexts.org/Courses/Bellarmine_University/BU%3A_Chem_103(Christianson)/ Phase_1%3A_Chemistry_Essentials/4%3A_Simple Chemical_Reactions/4.1%3A_Chemical_ Reaction_Equations ,May 2019.
[5] EMBOpress. Deep learning for computational biology. Christof Angermueller, Tanel Pärnamaa, Et al. Accessed at: http://msb.embopress.org/content/12/7/878 ,May 2019.
[6]Hu Zilonga, Tang Jinshan, Et al. Deep learning for image-based cancer detection and diagnosis, A survey. Accessed at: https://www.sciencedirect.com/science/article/abs/pii/S0031320318301845 , May 2019.
Contacts
If you want to keep updated with my latest articles and projects follow me on Medium and subscribe to my mailing list. These are some of my contacts details:
